Nat Med | Indlela ye-multi-omics yokwenza imephu ye-tumor edibeneyo, i-immune kunye ne-microbial landscape yomhlaza we-colorectal ibonisa intsebenziswano ye-microbiome kunye ne-immune system.
Nangona i-biomarkers yomhlaza we-colon ephambili ifundwe ngokubanzi kwiminyaka yakutshanje, izikhokelo zeklinikhi zangoku zixhomekeke kuphela kwi-tumor-lymph node-metastasis staging kunye nokufumanisa iziphene ze-DNA zokulungisa (MMR) okanye ukungazinzi kwe-microsatellite (MSI) (ukongeza kuvavanyo oluqhelekileyo lwe-pathology) ukumisela iingcebiso zonyango. Abaphandi baye baqaphela ukunqongophala kobudlelwane phakathi kweempendulo ze-gene-based immune-based response, iiprofayili ze-microbial, kunye ne-tumor stroma kwi-Cancer Genome Atlas (TCGA) ye-colorectal cancer cohort kunye nokusinda kwesigulana.
Njengoko uphando luye lwaqhubela phambili, iimpawu zobungakanani bomhlaza we-colorectal osisiseko, kubandakanya i-cell cell, i-immune, i-stromal, okanye i-microbial nature yomhlaza, ziye zaxelwa ukuba zihambelana kakhulu neziphumo zeklinikhi, kodwa kusekho ukuqonda okulinganiselweyo malunga nendlela ukusebenzisana kwabo okuchaphazela ngayo iziphumo zesigulana.
Ukusabalalisa ubudlelwane phakathi kobunzima be-phenotypic kunye nesiphumo, iqela labaphandi abavela kwiSidra Institute of Medical Research eQatar kutshanje iphuhlise kwaye iqinisekise inqaku elidibeneyo (mICRoScore) elichonga iqela lezigulane ezinemilinganiselo emihle yokuphila ngokudibanisa iimpawu ze-microbiome kunye ne-immune rejection constants (ICR). Iqela lenze uhlalutyo olubanzi lwe-genomic yeesampulu ezitsha ezinomkhenkce ezivela kwizigulana ezingama-348 ezinomhlaza ophambili we-colorectal, kubandakanya ulandelelwano lwe-RNA lwamathumba kunye nokudibanisa izicubu ezinempilo, ulandelelwano olupheleleyo lwe-exome, i-T-cell receptor enzulu kunye ne-16S yebhaktiriya ye-rRNA yolandelelwano yofuzo, eyongezelelwa yi-tumor genome eyongezelelweyo ukuqhubela phambili uphawu lwe-tumor genome. Uphononongo lwapapashwa kwiNature Medicine njenge "I-tumor edibeneyo, i-immune kunye ne-microbiome atlas ye-colon cancer".

Inqaku elipapashwe kwiNature Medicine
AC-ICAM Overview
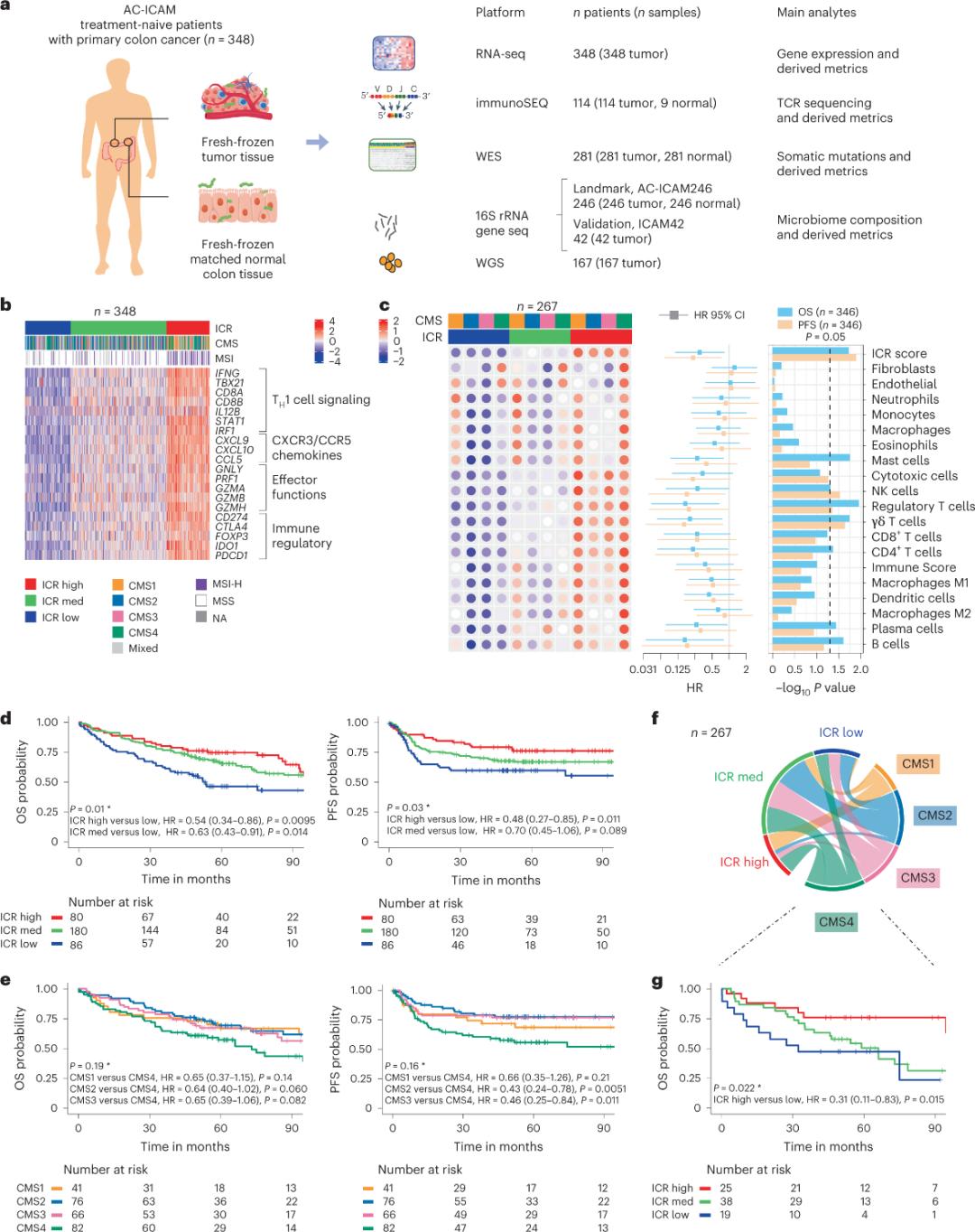
Abaphandi basebenzise iqonga le-orthogonal genomic ukuhlalutya iisampulu zethumba enomkhenkce kwaye zihambelane nethishu yekholoni esempilweni (izibini ezihlala ziqhuma) kwizigulana ezinokuxilongwa komhlaza wekoloni ngaphandle konyango lwenkqubo. Ngokusekelwe kwi-whole-exome sequencing (WES), i-RNA-seq yokulawula umgangatho wedatha, kunye nokuhlolwa kweekhrayitheriya zokubandakanywa, idatha ye-genomic evela kwizigulane ze-348 zigcinwe kwaye zisetyenziselwa uhlalutyo olusezantsi kunye nokulandelwa okuphakathi kweminyaka eyi-4.6. Iqela lophando libize lo vimba we-Sidra-LUMC AC-ICAM: Imephu kunye nesikhokelo sokusebenzisana kwe-immune-cancer-microbiome (Umfanekiso 1).
Ukuhlelwa kweemolekyuli kusetyenziswa i-ICR
Ukubamba iseti yeemodyuli zeempawu zofuzo zokuzikhusela kwisifo somhlaza esiqhubekayo, esibizwa ngokuba yi-immune constant rejection (ICR), iqela lophando liye layiphucula i-ICR ngokuyidibanisa kwiphaneli yemfuza engama-20 egubungela iindidi ezahlukeneyo zomhlaza, kubandakanya imelanoma, umhlaza wesinyi, kunye nomhlaza wamabele. I-ICR iye yadibaniswa nempendulo ye-immunotherapy kwiindidi ezahlukeneyo zomhlaza, kubandakanywa nomhlaza webele.
Okokuqala, abaphandi baqinisekisa isignesha ye-ICR ye-AC-ICAM cohort, usebenzisa i-ICR gene-based co-classification approach to classifying the cohort in three clusters / subtypes omzimba: i-ICR ephezulu (i-hot tumors), i-ICR ephakathi kunye ne-ICR ephantsi (iithumba ezibandayo) (Umfanekiso 1b). Abaphandi babonakalisa ukuthambekela kwe-immune ehambelana ne-subtypes ye-molecular subtypes (CMS), ulwahlulo olusekwe kwi-transcriptome yomhlaza wekoloni. iindidi zeCMS zibandakanya iCMS1/immune, CMS2/canonical, CMS3/metabolic and CMS4/mesenchymal. Uhlalutyo lubonise ukuba amanqaku e-ICR anxulunyaniswa kakubi neendlela ezithile zeeseli zomhlaza kuzo zonke ii-subtypes ze-CMS, kwaye ulungelelwaniso olulungileyo kunye neendlela ezinxulumene ne-immunosuppressive kunye ne-stromal-zabonwa kuphela kwii-CMS4 tumors.
Kuzo zonke i-CMS, ubuninzi be-cell killer (NK) kunye ne-T cell subsets yayiphezulu kwi-ICR ephezulu ye-immune subtypes, kunye nokuhlukahluka okukhulu kwezinye iiseti ze-leukocyte (Umfanekiso 1c) .I-ICR i-immune subtypes ine-OS eyahlukileyo kunye ne-PFS, kunye nokunyuka okuqhubekayo kwi-ICR ukusuka kwi-low ukuya phezulu (Figure 1d), eqinisekisa indima ye-ICR yombala we-prognostic yendima yomhlaza.
Umzobo 1. Uyilo lokufunda lwe-AC-ICAM, isignesha ye-gene enxulumene nomzimba, i-immune kunye ne-molecular subtypes kunye nokusinda.
I-ICR ibamba i-tumor-eriched, i-clonal amplified T cells
Kuphela ligcuntswana iiseli T ingena kwizicubu ithumba ziye zaxelwa ukuba ngqo antigens ithumba (ngaphantsi kwe-10%). Ke ngoko, uninzi lweeseli ze-T ze-intra-tumor zibizwa ngokuba ziiseli T ezibukeleyo (iiseli T ezibukeleyo). Ukulungelelaniswa okunamandla kunye nenani leeseli ze-T eziqhelekileyo ezine-TCRs ezivelisayo zabonwa kwi-stromal cell kunye ne-leukocyte subpopulations (efunyenwe yi-RNA-seq), engasetyenziselwa ukuqikelela i-T cell subpopulations (Umfanekiso 2a). Kumaqela e-ICR (ngokubanzi kunye nokuhlelwa kwe-CMS), i-clonality ephezulu ye-immune SEQ TCRs yabonwa kwi-ICR-high kunye ne-CMS subtype CMS1 / amaqela omzimba (Umfanekiso 2c), kunye neyona ndawo iphezulu ye-ICR-high tumors. Ukusebenzisa i-transcriptome yonke (i-18,270 genes), i-ICR i-genenes ezintandathu (i-IFNG, i-STAT1, i-IRF1, i-CCL5, i-GZMA, kunye ne-CXCL10) yayiphakathi kweentlobo ezilishumi eziphezulu ezinxulumene ne-TCR immune SEQ clonality (Figure 2d). I-ImmunoSEQ TCR clonality idityaniswe ngamandla kuninzi lwe-ICR yofuzo kunolungelelwaniso olujongwe kusetyenziswa iziphawuli ze-CD8+ eziphendula ithumba (Umfanekiso 2f kunye ne-2g). Ukuqukumbela, olu hlalutyo lungentla lucebisa ukuba utyikityo lwe-ICR lubamba ubukho beeseli ze-T ezityebileyo, ezikhuliswe ngokudibeneyo kwaye zinokucacisa iziphumo zayo.
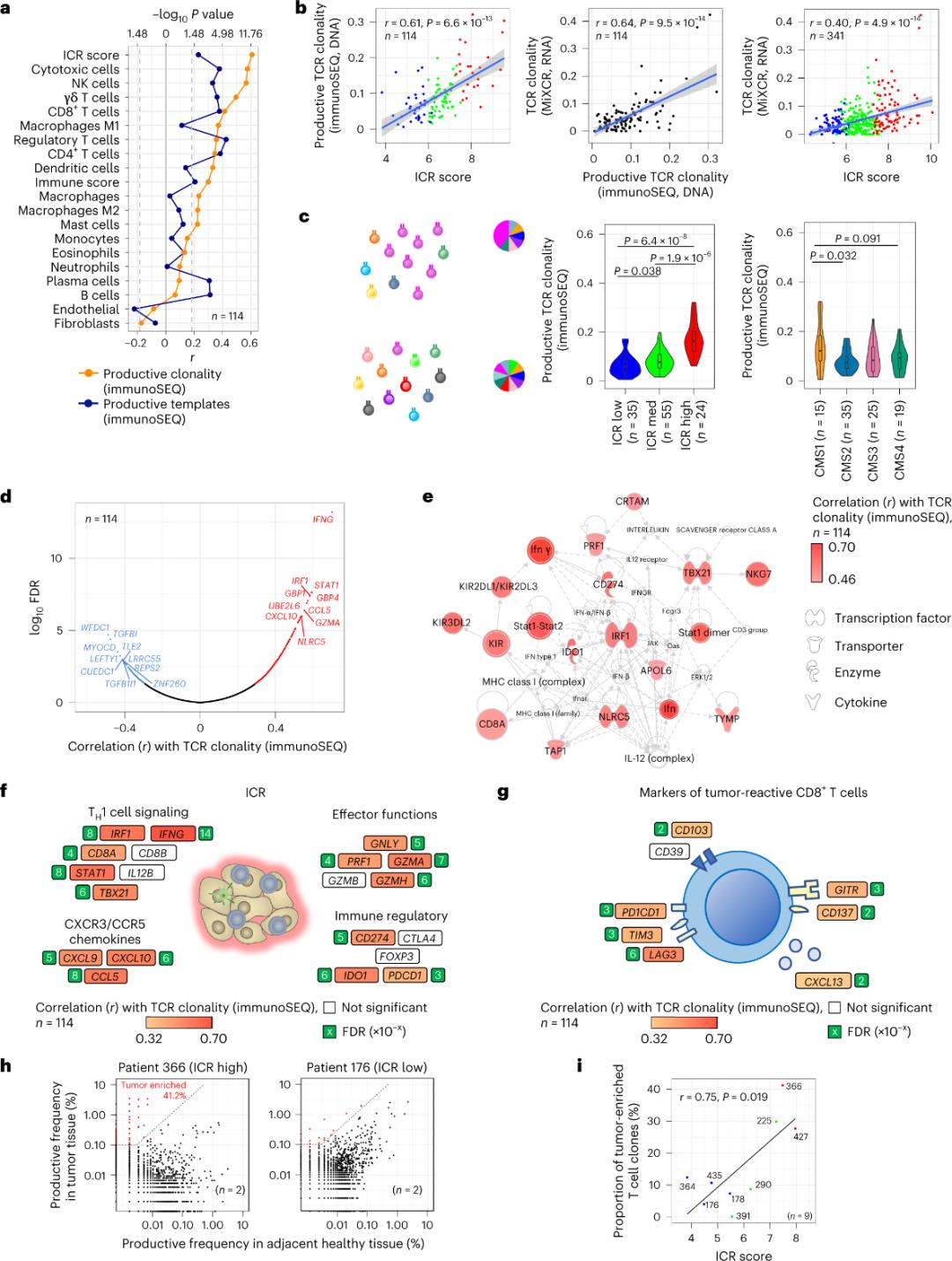
Umzobo 2. Iimethrikhi ze-TCR kunye nokulungelelaniswa kunye neegenes ezinxulumene ne-immune, i-immune kunye ne-molecular subtypes.
Ukubunjwa kwe-Microbiome kwizicubu zomhlaza we-colon onempilo
Abaphandi benza i-16S rRNA ngokulandelelana usebenzisa i-DNA ekhutshwe kwi-tumor ehambelanayo kunye ne-colon tissue enempilo evela kwizigulane ze-246 (Umfanekiso 3a). Ukuqinisekisa, abaphandi bahlalutya ngakumbi i-16S rRNA idatha yolandelelwano lwemfuza ukusuka kwisampulu eyongezelelweyo ye-42 ye-tumor engazange ihambelane ne-DNA eqhelekileyo ekhoyo ukuze ihlalutywe. Okokuqala, abaphandi bathelekisa ubuninzi bezityalo phakathi kwamathumba ahambelanayo kunye nezicubu zekholoni ezisempilweni. I-Clostridium perfringens yanda kakhulu kwii-tumors xa kuthelekiswa neesampuli eziphilileyo (Umfanekiso 3a-3d). Kwakungekho mahluko ubalulekileyo kwiiyantlukwano ze-alpha (ukwahlukana kunye nobuninzi beentlobo kwisampulu enye) phakathi kwe-tumor kunye neesampuli eziphilileyo, kunye nokunciphisa okuthobekileyo kwi-microbial diversity kwabonwa kwi-ICR-high tumors ngokumalunga ne-ICR-low tumors.
Ukubona unxulumano olufanelekileyo lweklinikhi phakathi kweeprofayili ze-microbial kunye neziphumo zeklinikhi, abaphandi bajonge ukusebenzisa i-16S rRNA idatha yokulandelelana kofuzo ukuchonga iimpawu ze-microbiome eziqikelela ukusinda. Kwi-AC-ICAM246, abaphandi baqhuba imodeli yokubuyisela i-OS Cox ekhethiweyo iimpawu ze-41 kunye ne-non-zero coefficients (ehambelana nomngcipheko wokufa okungafaniyo), ebizwa ngokuba yi-MBR classifiers (Figure 3f).
Kulo qela loqeqesho (i-ICAM246), inqaku eliphantsi le-MBR (MBR <0, i-MBR ephantsi) lidibene nomngcipheko ophantsi wokufa (85%). Abaphandi baqinisekisile umbutho phakathi kwe-MBR ephantsi (umngcipheko) kunye ne-OS ende kwii-cohorts ezimbini ezizimeleyo eziqinisekisiweyo (i-ICAM42 kunye ne-TCGA-COAD). (Umfanekiso 3) Uphononongo lubonise ukulungelelaniswa okuqinileyo phakathi kwe-cocci endogastric kunye namanqaku e-MBR, afana ne-tumor kunye ne-colon tissue enempilo.
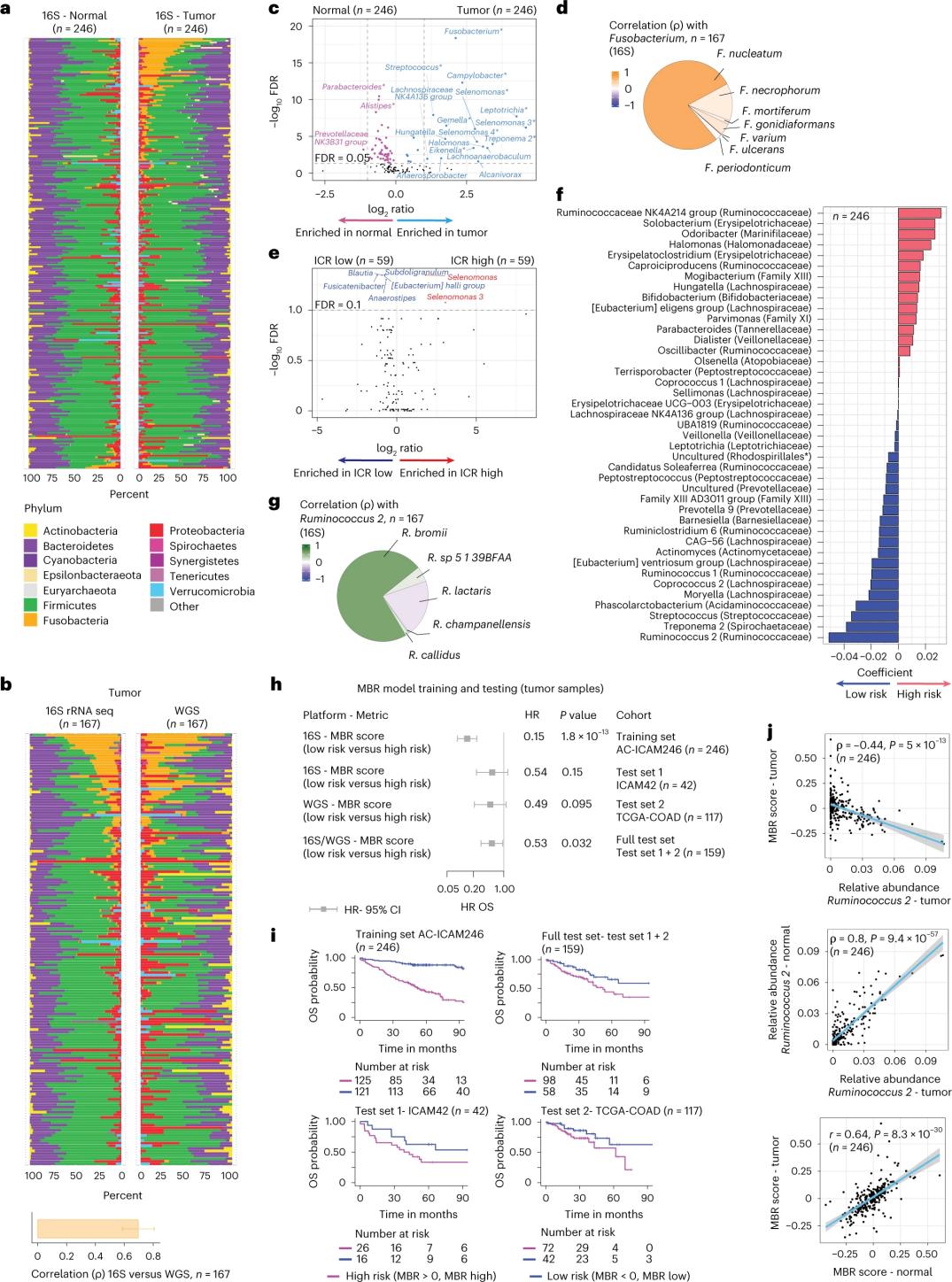
Umzobo 3. I-Microbiome kwi-tumor kunye nezicubu ezinempilo kunye nobudlelwane kunye ne-ICR kunye nokusinda kwesigulane.
Ukuqukumbela
Indlela ye-multi-omics esetyenzisiweyo kolu phononongo yenza ukuba kubonwe ngokucokisekileyo kunye nohlalutyo lwesiginesha yemolekyuli yempendulo ye-immune kumhlaza we-colorectal kwaye ityhila intsebenziswano phakathi kwe-microbiome kunye ne-immune system. Ulandelelwano olunzulu lwe-TCR yethumba kunye nezihlunu ezisempilweni zibonise ukuba isiphumo se-ICR sinokubangelwa kukukwazi ukubamba i-tumor-etyetyiswe kwaye mhlawumbi i-tumor antigen-specific cell clones ye-T.
Ngokuhlalutya ukubunjwa kwe-tumor microbiome usebenzisa i-16S rRNA gene sequencing kwiisampuli ze-AC-ICAM, iqela lichonge isiginitsha ye-microbiome (inqaku lomngcipheko we-MBR) kunye nexabiso elinamandla le-prognostic. Nangona olu tyikityo luthatyathwe kwiisampulu zethumba, bekukho ulungelelwaniso olomeleleyo phakathi kwe-colorectum esempilweni kunye nenqanaba lomngcipheko we-tumor MBR, ecebisa ukuba olu tyikityo lunokuthi lubambe ukubunjwa kwe-gut microbiome yezigulana. Ngokudibanisa amanqaku e-ICR kunye ne-MBR, kwakunokwenzeka ukuchonga kunye nokuqinisekisa i-biomarker yomfundi we-multi-omic eqikelela ukusinda kwizigulane ezinomhlaza wekoloni. Iseti yedatha yophononongo ye-multi-omic ibonelela ngesixhobo sokuqonda ngcono ibhayoloji yomhlaza wekolon kunye nokunceda ukufumanisa iindlela zonyango zomntu.
Ixesha lokuposa: Jun-15-2023
 中文网站
中文网站